#SeuratData::InstallData("celegans.embryo")
obj <- LoadData("celegans.embryo", type="default")
obj[["percent.mt"]] <- PercentageFeatureSet(obj, pattern = "^MT-")
obj <- subset(obj, subset = nFeature_RNA > 200 & nFeature_RNA < 2500 & percent.mt < 5)
obj <- NormalizeData(obj)
obj <- FindVariableFeatures(obj, selection.method = "vst", nfeatures = 2000)
all.genes <- rownames(obj)
obj <- ScaleData(obj, features = all.genes)
obj <- RunPCA(obj, features = VariableFeatures(object = obj))
obj = RunUMAP(object = obj, dims = 1:10)
# Example usage
#entropy <- shannon_entropy(obj, method="predefined")
entropy <- shannon_entropy(obj, method="RCSA")
## Calculating reference entropy...
##
1: Stablility = 0.000006
## Calculating reference entropy...
##
2: Stablility = 0.000000
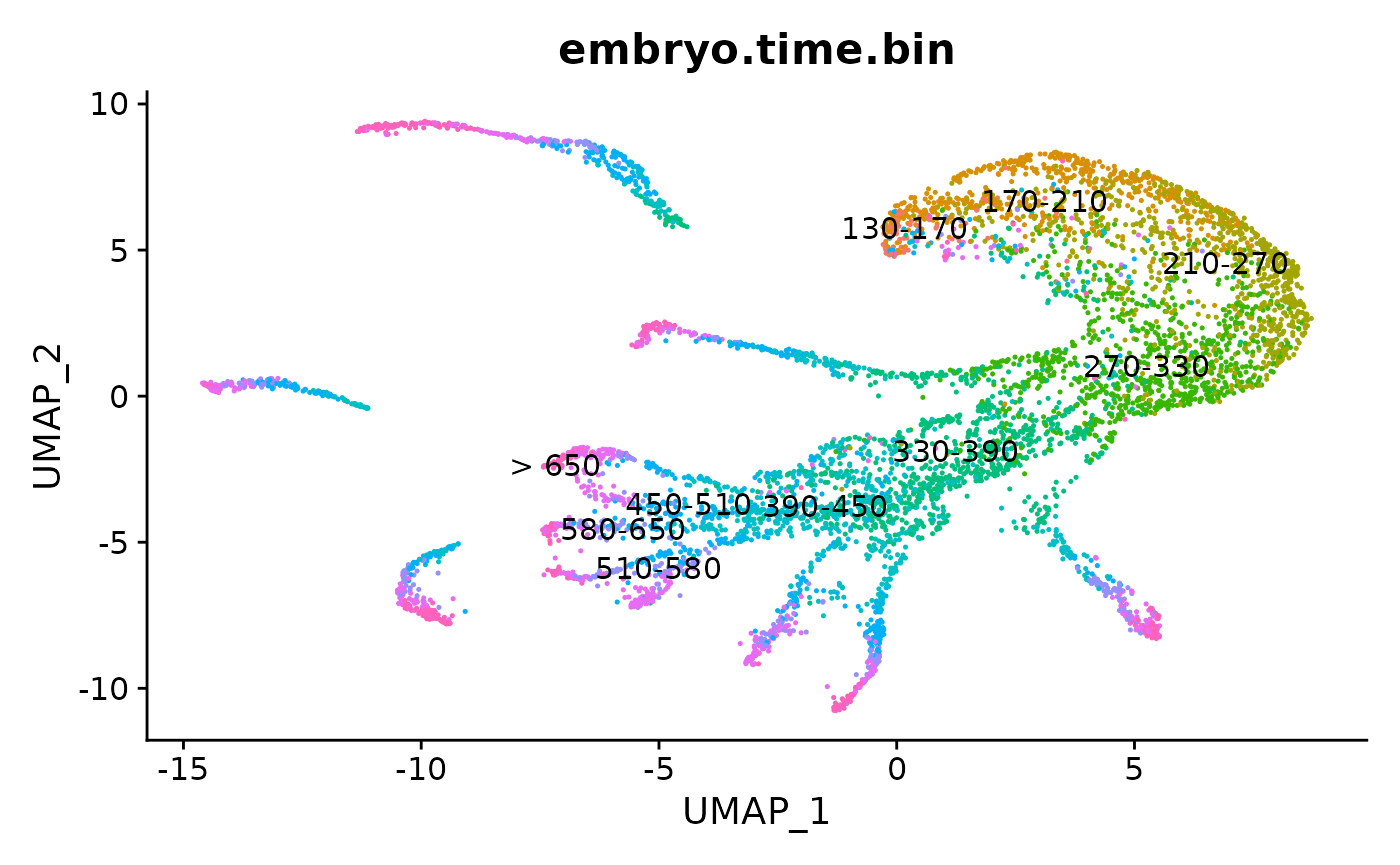
DimPlot(obj,reduction = "umap", group.by = "embryo.time.bin", label = T) + NoLegend()
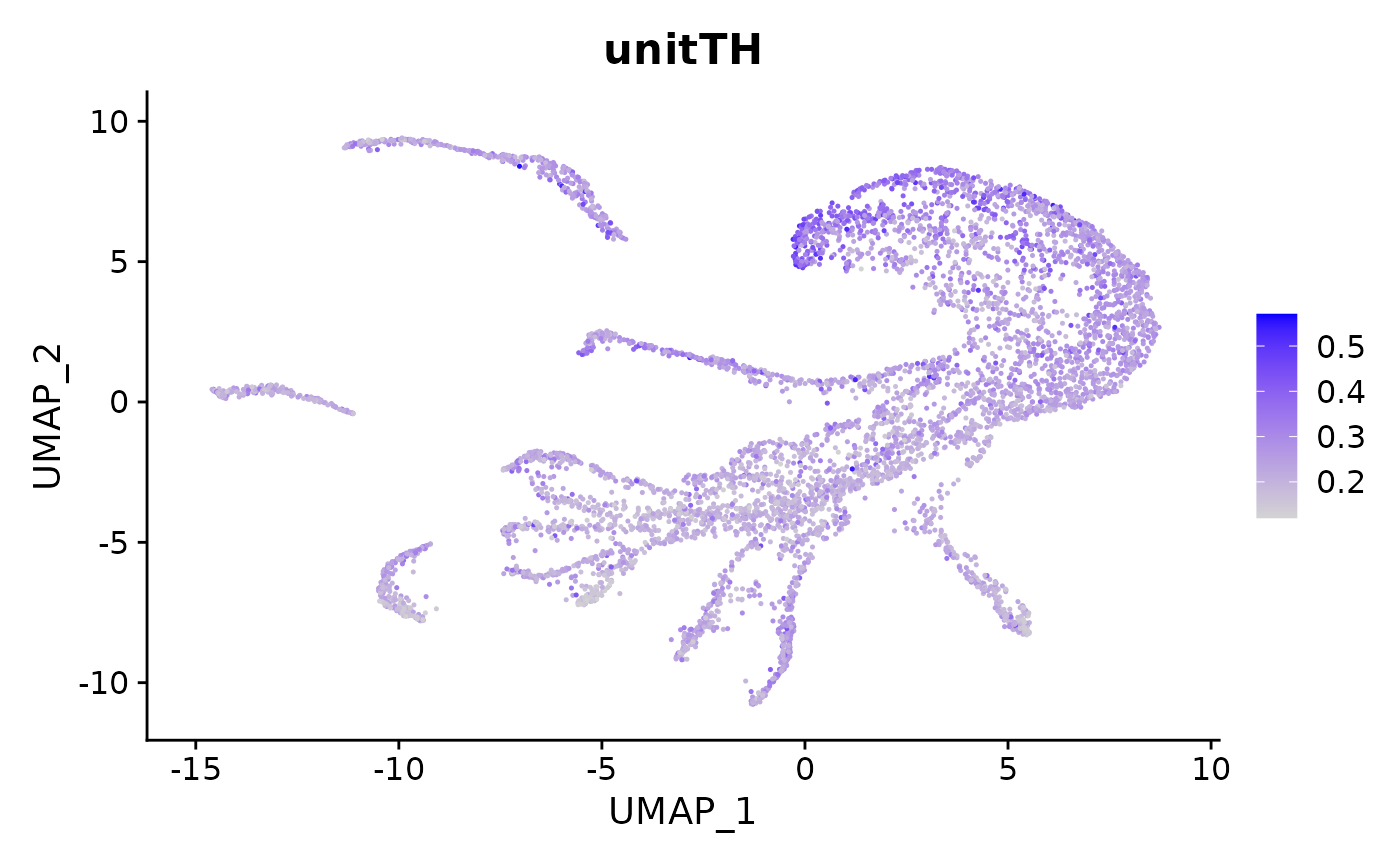
#SeuratData::InstallData("pbmc3k")
pbmc3k.final <- LoadData("pbmc3k", type = "pbmc3k.final")
# Example usage
#entropy <- shannon_entropy(pbmc3k.final, method="predefined")
entropy <- shannon_entropy(pbmc3k.final, method="RCSA")
## Calculating reference entropy...
##
1: Stablility = 0.000024
## Calculating reference entropy...
##
2: Stablility = 0.000001
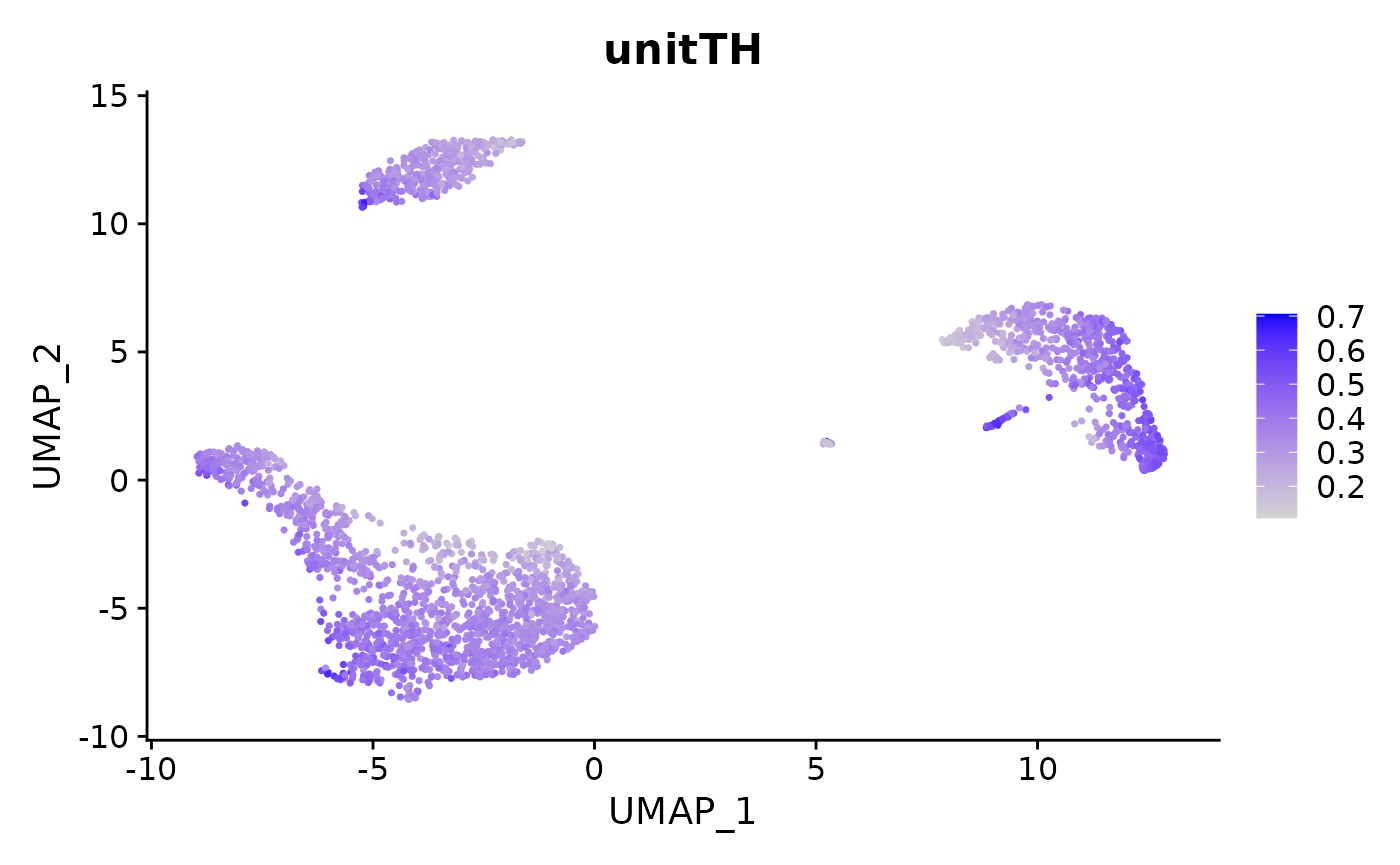
pbmc3k.final = AddMetaData(pbmc3k.final, entropy, col.name = "unitTH")
#summary(pbmc_small@meta.data)
#colnames(pbmc3k.final@meta.data)
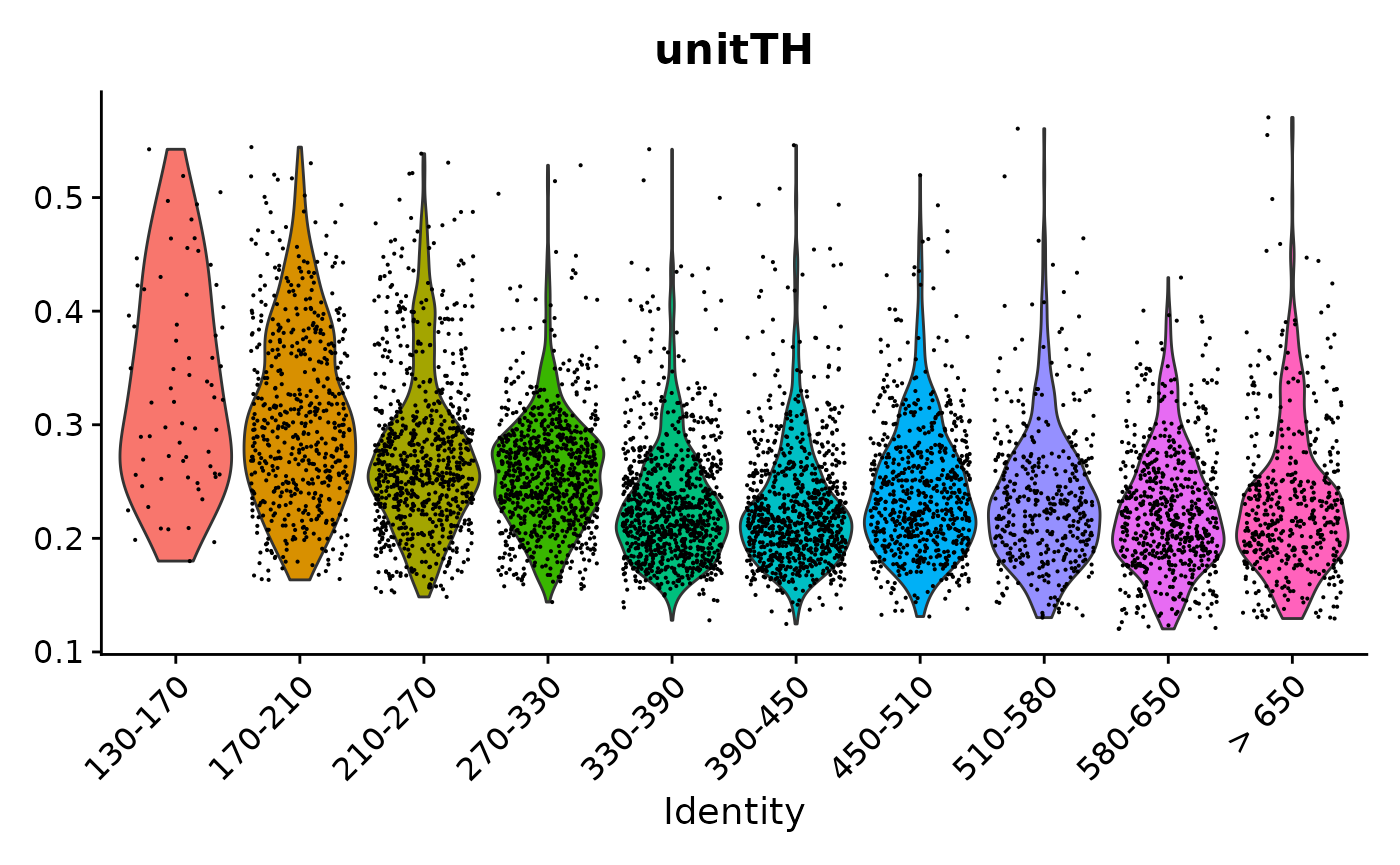
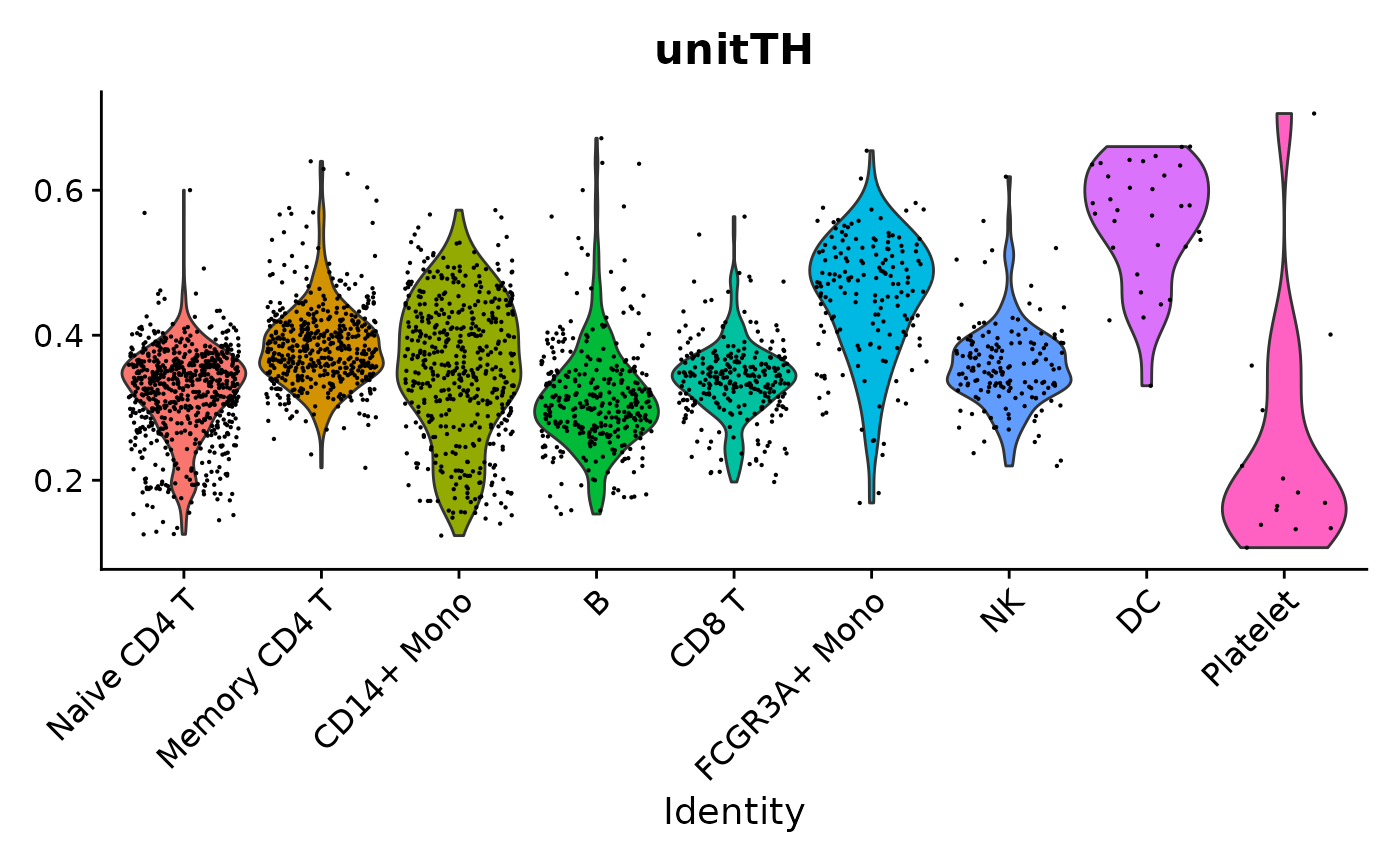
VlnPlot(pbmc3k.final, features = "unitTH", group.by = "seurat_annotations") + NoLegend()
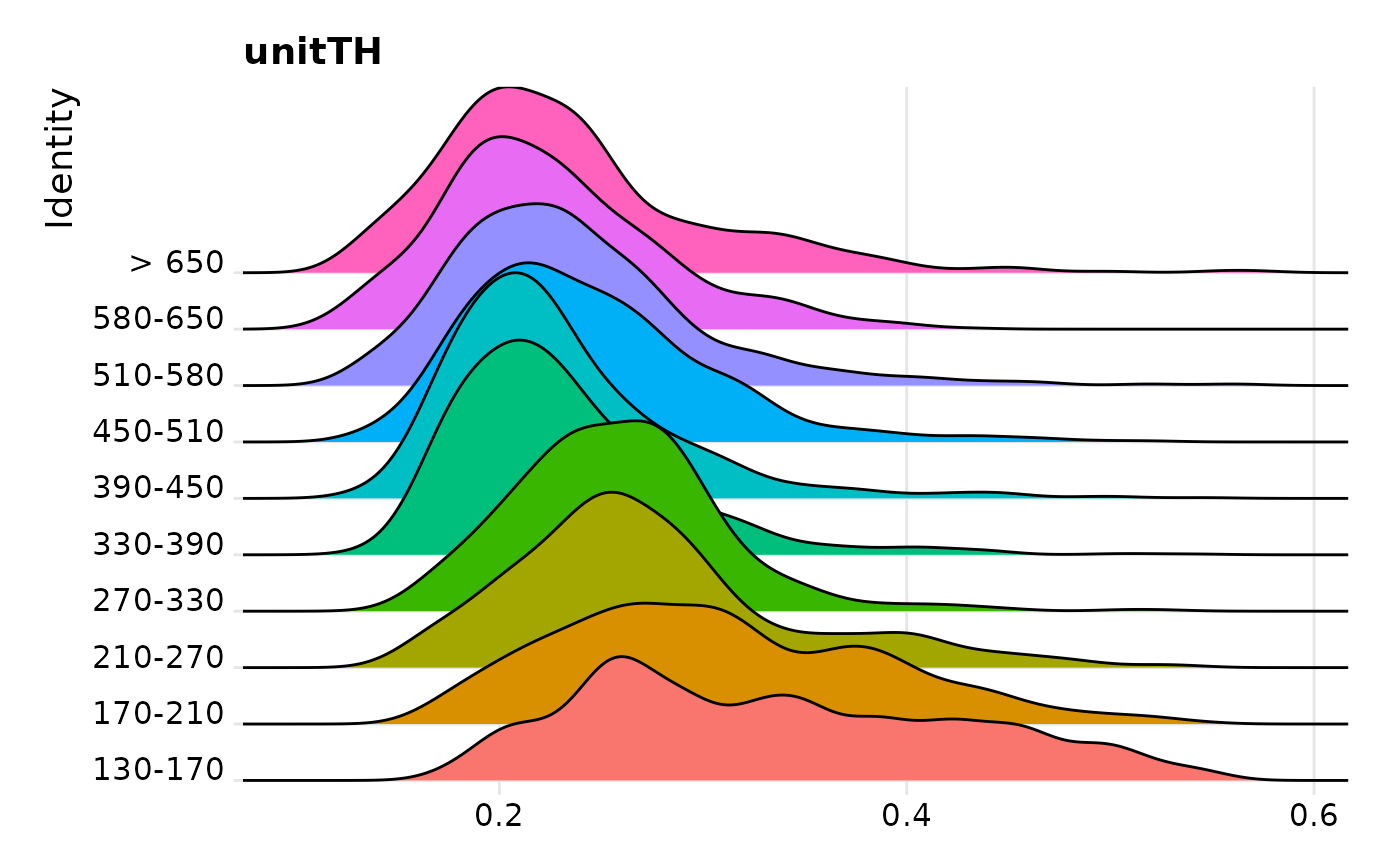
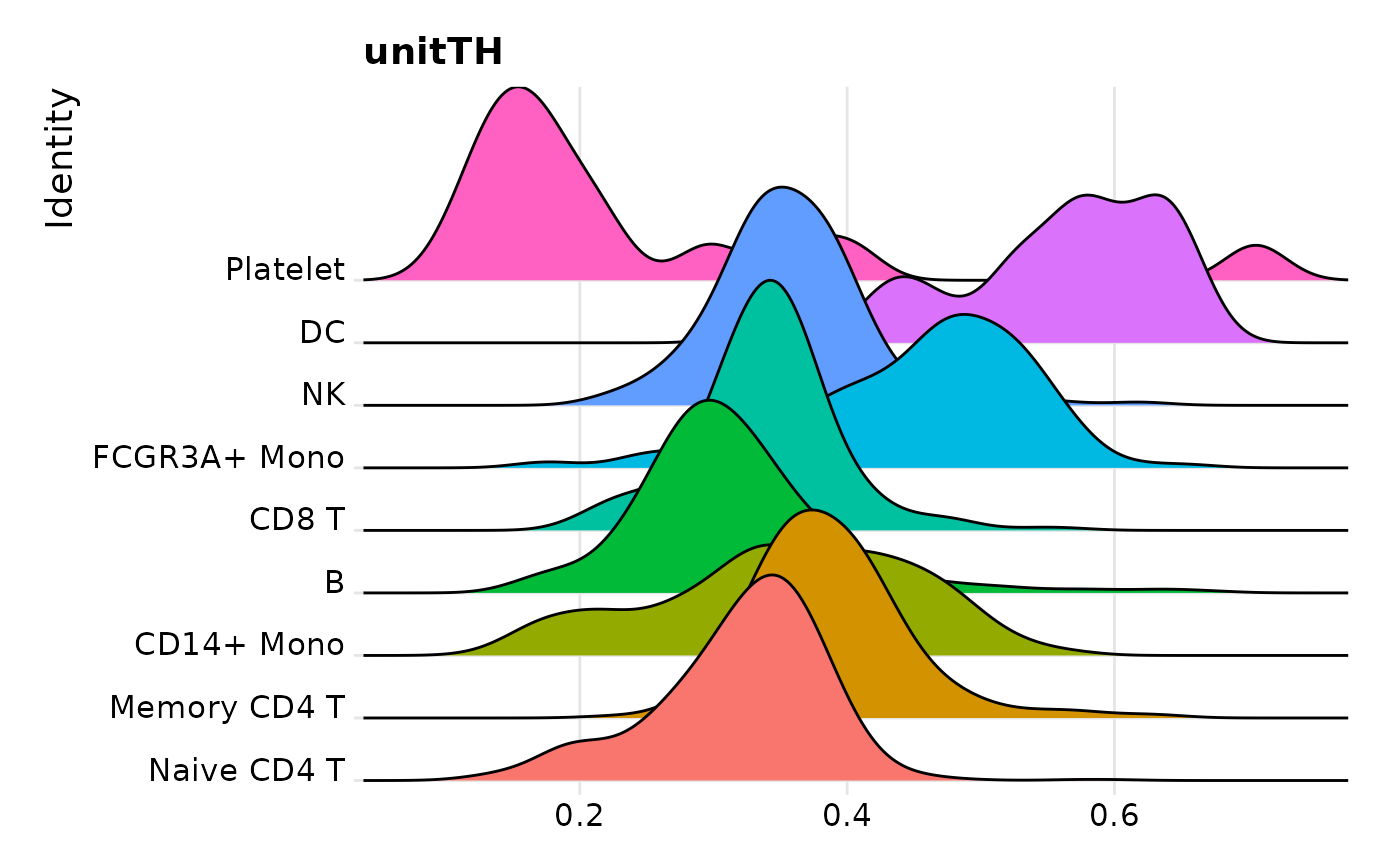
RidgePlot(pbmc3k.final, features = "unitTH", group.by = "seurat_annotations") + NoLegend()
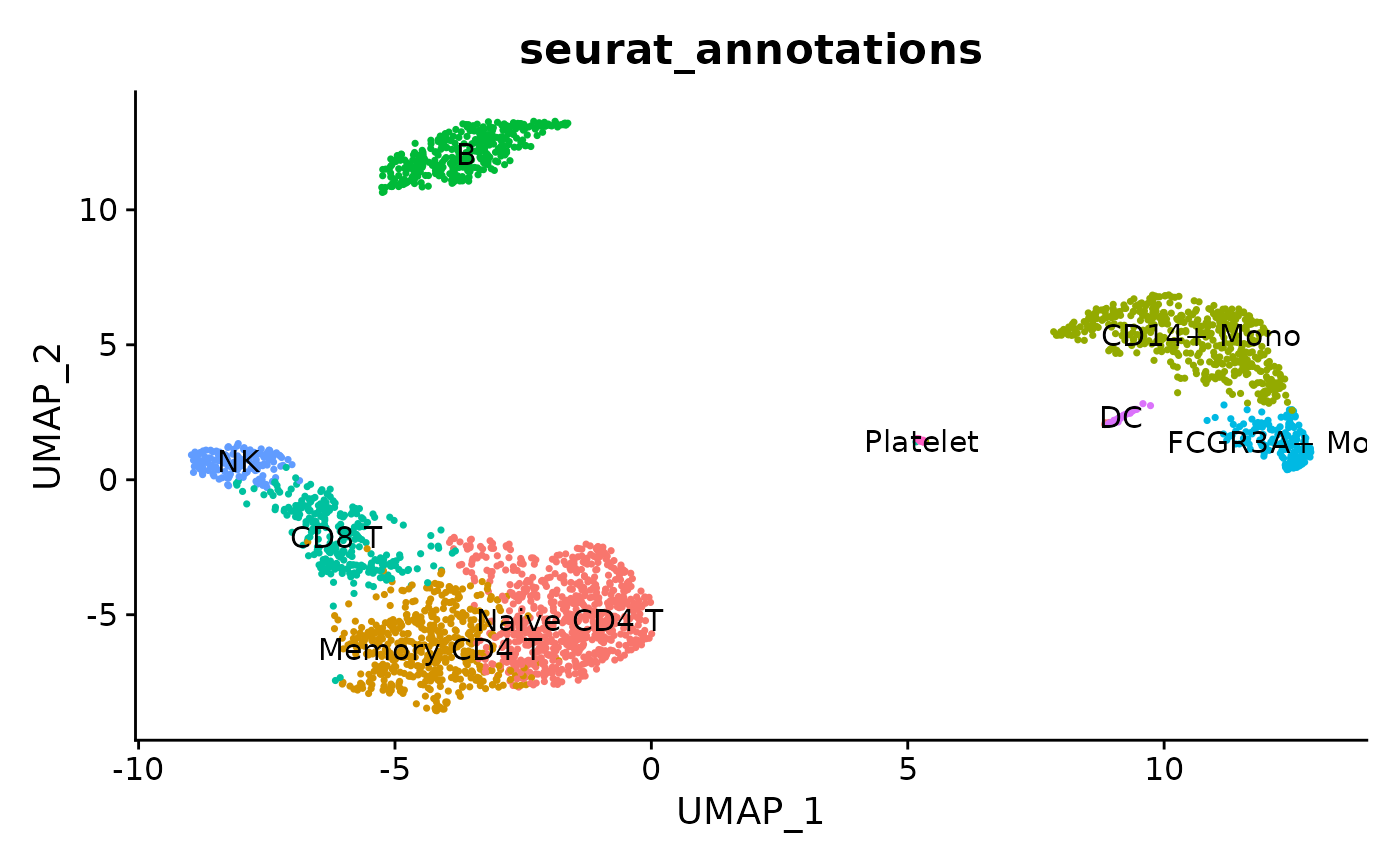
DimPlot(pbmc3k.final,reduction = "umap", group.by = "seurat_annotations", label = T) + NoLegend()