##
## clusterProfiler v3.99.2 For help: https://guangchuangyu.github.io/software/clusterProfiler
##
## If you use clusterProfiler in published research, please cite:
## Guangchuang Yu, Li-Gen Wang, Yanyan Han, Qing-Yu He. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology. 2012, 16(5):284-287.
##
## Attaching package: 'clusterProfiler'
## The following object is masked from 'package:stats':
##
## filter
## Loading required package: AnnotationDbi
## Loading required package: stats4
## Loading required package: BiocGenerics
## Loading required package: parallel
##
## Attaching package: 'BiocGenerics'
## The following objects are masked from 'package:parallel':
##
## clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,
## clusterExport, clusterMap, parApply, parCapply, parLapply,
## parLapplyLB, parRapply, parSapply, parSapplyLB
## The following objects are masked from 'package:stats':
##
## IQR, mad, sd, var, xtabs
## The following objects are masked from 'package:base':
##
## anyDuplicated, append, as.data.frame, basename, cbind, colnames,
## dirname, do.call, duplicated, eval, evalq, Filter, Find, get, grep,
## grepl, intersect, is.unsorted, lapply, Map, mapply, match, mget,
## order, paste, pmax, pmax.int, pmin, pmin.int, Position, rank,
## rbind, Reduce, rownames, sapply, setdiff, sort, table, tapply,
## union, unique, unsplit, which.max, which.min
## Loading required package: Biobase
## Welcome to Bioconductor
##
## Vignettes contain introductory material; view with
## 'browseVignettes()'. To cite Bioconductor, see
## 'citation("Biobase")', and for packages 'citation("pkgname")'.
## Loading required package: IRanges
## Loading required package: S4Vectors
##
## Attaching package: 'S4Vectors'
## The following object is masked from 'package:clusterProfiler':
##
## rename
## The following objects are masked from 'package:base':
##
## expand.grid, I, unname
##
## Attaching package: 'IRanges'
## The following object is masked from 'package:clusterProfiler':
##
## slice
##
## Attaching package: 'AnnotationDbi'
## The following object is masked from 'package:clusterProfiler':
##
## select
##
library(ggplot2)
data(geneList, package="DOSE")
gene <- names(geneList)[abs(geneList) > 2]
gene.df <- bitr(gene, fromType = "ENTREZID",
toType = c("ENSEMBL", "SYMBOL"),
OrgDb = org.Hs.eg.db)
## 'select()' returned 1:many mapping between keys and columns
## Warning in bitr(gene, fromType = "ENTREZID", toType = c("ENSEMBL", "SYMBOL"), :
## 0.48% of input gene IDs are fail to map...
## ENTREZID ENSEMBL SYMBOL
## 1 4312 ENSG00000196611 MMP1
## 2 8318 ENSG00000093009 CDC45
## 3 10874 ENSG00000109255 NMU
## 4 55143 ENSG00000134690 CDCA8
## 5 55388 ENSG00000065328 MCM10
## 6 991 ENSG00000117399 CDC20
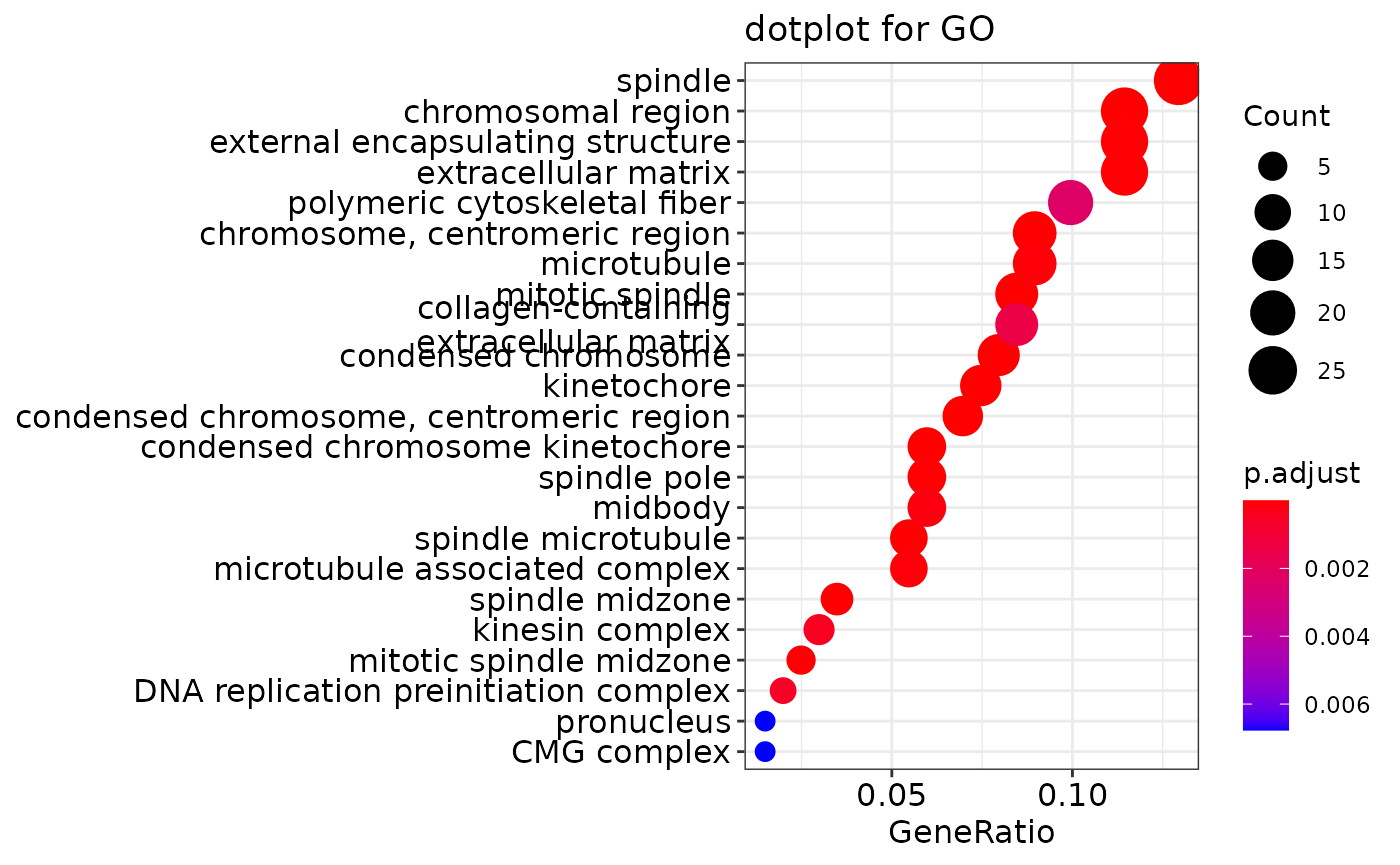
ego <- enrichGO(gene = gene,
universe = names(geneList),
OrgDb = org.Hs.eg.db,
ont = "CC",
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05,
readable = TRUE)
head(ego)
## ID Description GeneRatio
## GO:0005819 GO:0005819 spindle 26/201
## GO:0072686 GO:0072686 mitotic spindle 17/201
## GO:0000775 GO:0000775 chromosome, centromeric region 18/201
## GO:0098687 GO:0098687 chromosomal region 23/201
## GO:0000779 GO:0000779 condensed chromosome, centromeric region 14/201
## GO:0000776 GO:0000776 kinetochore 15/201
## BgRatio pvalue p.adjust qvalue
## GO:0005819 299/11840 6.490593e-12 1.908234e-09 1.735380e-09
## GO:0072686 129/11840 5.480692e-11 8.056617e-09 7.326819e-09
## GO:0000775 156/11840 1.385677e-10 1.206452e-08 1.097168e-08
## GO:0098687 269/11840 1.641432e-10 1.206452e-08 1.097168e-08
## GO:0000779 90/11840 3.056082e-10 1.796976e-08 1.634199e-08
## GO:0000776 109/11840 4.155863e-10 2.036373e-08 1.851911e-08
## geneID
## GO:0005819 CDCA8/CDC20/KIF23/CENPE/ASPM/DLGAP5/SKA1/NUSAP1/TPX2/TACC3/NEK2/CDK1/MAD2L1/KIF18A/BIRC5/KIF11/TRAT1/TTK/AURKB/PRC1/KIFC1/KIF18B/KIF20A/AURKA/CCNB1/KIF4A
## GO:0072686 KIF23/CENPE/ASPM/SKA1/NUSAP1/TPX2/TACC3/CDK1/MAD2L1/KIF18A/KIF11/TRAT1/AURKB/PRC1/KIFC1/KIF18B/AURKA
## GO:0000775 CDCA8/CENPE/NDC80/TOP2A/HJURP/SKA1/NEK2/CENPM/CENPN/ERCC6L/MAD2L1/KIF18A/CDT1/BIRC5/TTK/NCAPG/AURKB/CCNB1
## GO:0098687 CDCA8/CENPE/NDC80/TOP2A/HJURP/SKA1/NEK2/CENPM/RAD51AP1/CENPN/CDK1/ERCC6L/MAD2L1/KIF18A/CDT1/BIRC5/EZH2/TTK/NCAPG/AURKB/CHEK1/CCNB1/MCM5
## GO:0000779 CENPE/NDC80/HJURP/SKA1/NEK2/CENPM/CENPN/ERCC6L/MAD2L1/CDT1/BIRC5/NCAPG/AURKB/CCNB1
## GO:0000776 CENPE/NDC80/HJURP/SKA1/NEK2/CENPM/CENPN/ERCC6L/MAD2L1/KIF18A/CDT1/BIRC5/TTK/AURKB/CCNB1
## Count
## GO:0005819 26
## GO:0072686 17
## GO:0000775 18
## GO:0098687 23
## GO:0000779 14
## GO:0000776 15